Introduction
There are a broad range of issues relevant to the question of altering milk composition. A general review of all the issues is beyond the scope of this paper. The paper will address only the theoretical and practical aspects of the topic that are relevant from a research perspective using DNA technology. Our purpose is to explain how it is now possible to use DNA technology to exploit existing genetic variation in the dairy cattle population and thereby alter milk composition to benefit the dairy industry.DNA technology includes an extensive scope of research techniques used to study the molecular biology of cells and organisms (14). Although the techniques can be applied to create transgenic animals, the use of that approach to alter milk composition has been extremely limited. On the other hand, the mammary gland has been a target of transgenic methods with the aim of producing high value human pharmaceuticals (5). Despite the success in that area, the current lack of understanding of the complexities of multigene interactions has limited progress on transgenic applications in animal agriculture in general. In contrast, our research strategy uses the power of DNA technology, but relies on the wisdom of nature to achieve its ultimate objective.
Our research strategy involves identifying existing genes that have naturally evolved as regulators of milk synthesis and therefore coadapted with all other genes. The eloquent complexity of proper gene function in cows living productively in a dairy environment is largely due to the process of natural selection.
The conceptual basis for our research in the area of altering milk composition using DNA technology is relatively simple. We believe that gene expression in mammary cells lies at the heart of the mechanisms controlling milk synthesis and, therefore, milk composition. After all, it is known that the genes of a cell carry all the information required to specify the structure and function of the cell. In essence, if a gene in the mammary gland is not turned on (expressed), then the protein encoded by the gene will also not be found in the mammary gland. Similarly, if the expression of a certain gene is increased in the mammary gland of cow A compared to cow B, then there will be more of the protein (encoded by the gene) in the mammary gland of cow A.
Important Questions
The concept that gene expression is related to milk composition raises two extremely important questions:Question 1: Can the genes that control milk composition be found?
The simple answer to the first question is yes. We now have the scientific ability, using DNA technology, to find all of the genes involved in regulating the synthesis of the components in milk. That task may seem impossible, especially considering that about 15,000 genes are expressed simultaneously in each mammary cell. However, as you will see by later comments in the Methods section, we are now applying a new and powerful molecular technique called differential display that allows us for the first time to identify the genes controlling milk synthesis. In other words, we finally at least have access to all the pieces to the lactation puzzle.
Question 2: If and when such genes are found can that knowledge be applied to benefit the dairy industry, and if so, how?
The simple answer to this second question is that the identification of the genes controlling milk synthesis would represent a landmark advance in dairy cattle breeding. The importance of such new knowledge about the bovine genome to the dairy industry cannot be emphasized enough. Indeed, steps to overcome the practical problems associated with transferring genotyping technology to the dairy industry must get underway now. To that end, we have developed simple and inexpensive genotyping methods for two important milk protein genes, namely k-casein (k-CN) and b-lactoglobulin (b-LG). The methods are now ready for implementation in the commercial dairy arena. As you will see by the description in the Methods section below, our genotyping procedure can also be used for any gene in any cow, regardless of sex or age.
Methods
Differential DisplayThis method is used to identify presently unknown genes that play key roles in regulating the synthesis of milk components.
Even though some of the most important advances in DNA technology were made over 20 years ago (1), and new methods continue to be developed and improved, the search for genes associated with dairy production traits of economic importance has been difficult, and in practical terms unproductive. In dairy genetics those elusive genes are commonly referred to as quantitative trait loci (QTL). Over ten years ago there was widespread excitement about using a technique called restriction fragment length polymorphism (RFLP) analysis for the genetic improvement of livestock. Although the RFLP technique was a major advance for molecular genetics, allowing us to identify differences in known genes, its use was limited because there were no known QTL to evaluate.
 The real research challenge has therefore been to identify QTL. Over the last decade there has been
renewed hope that QTL may be found as a result of the discovery of repetitive and potentially
informative regions in DNA called variable number of tandem repeats (VNTR), minisatellites,
and microsatellites. The repeat units themselves are not QTL, but at least in theory it should be
possible to map QTL by evaluating the repeat units in large-scale linkage studies (16). Although
the use of that strategy to find QTL will undoubtedly continue, it is likely that the use of
differential display will expand rapidly in a rush to find genes of economic importance.
The real research challenge has therefore been to identify QTL. Over the last decade there has been
renewed hope that QTL may be found as a result of the discovery of repetitive and potentially
informative regions in DNA called variable number of tandem repeats (VNTR), minisatellites,
and microsatellites. The repeat units themselves are not QTL, but at least in theory it should be
possible to map QTL by evaluating the repeat units in large-scale linkage studies (16). Although
the use of that strategy to find QTL will undoubtedly continue, it is likely that the use of
differential display will expand rapidly in a rush to find genes of economic importance.
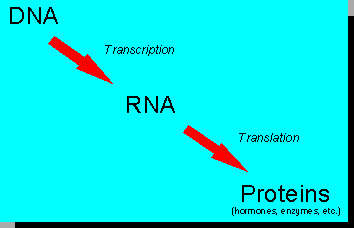
In order to demonstrate the utility of differential display for identifying genes involved in regulating milk synthesis we used growth hormone (GH) injection as a means to stimulate milk synthesis. To begin the differential display procedure we isolated messenger RNA from the mammary gland of a GH-treated cow and an untreated cow. Messenger RNA, as its name implies, is the molecule that carries the message encoded in DNA to where it is needed for building proteins (e.g., hormones, enzymes, etc.); proteins are the tools of cell function. That process of information transfer can be schematically depicted:
The differential display procedure itself is mainly based on the use of specially designed sets of short pieces of DNA (called primers) together with a technique called the polymerase chain reaction (PCR). The theoretical and methodological aspects of differential display have been reviewed (2). Figure 1 is a schematic representation of the main principles underlying the differential display procedure.
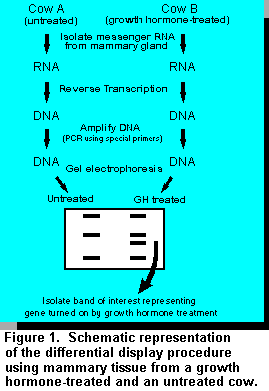 The first step in the differential display procedure simply involves reversing the natural "DNA to
RNA" process by transforming the RNA (isolated from the mammary gland) back into DNA. The
reason for using DNA is two-fold. First, it is easier to work with DNA than RNA; and second,
DNA is needed for the next step, which involves amplifying the DNA using the polymerase chain
reaction (Figure 1). After amplifying the DNA, we finally end up with a collection of DNA
molecules that actually represent only the mammary genes that are turned on (expressed). Therein
lies the "magic" of the differential display technique. The amplified DNA, representing only
expressed genes, is displayed on a gel to compare the genes expressed in a normal mammary
gland to those expressed in a mammary gland from a GH-treated cow.
The first step in the differential display procedure simply involves reversing the natural "DNA to
RNA" process by transforming the RNA (isolated from the mammary gland) back into DNA. The
reason for using DNA is two-fold. First, it is easier to work with DNA than RNA; and second,
DNA is needed for the next step, which involves amplifying the DNA using the polymerase chain
reaction (Figure 1). After amplifying the DNA, we finally end up with a collection of DNA
molecules that actually represent only the mammary genes that are turned on (expressed). Therein
lies the "magic" of the differential display technique. The amplified DNA, representing only
expressed genes, is displayed on a gel to compare the genes expressed in a normal mammary
gland to those expressed in a mammary gland from a GH-treated cow.
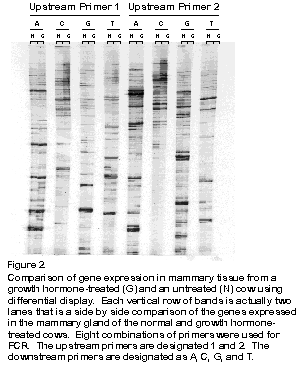
The usefulness and power of the differential display method is easily recognized upon examining a simultaneous comparison of the expression status of just a fraction (10%) of all 15,000 genes in the mammary gland of a GH-treated and untreated cow (Figure 2); compare the black bands that are side by side in each pair of vertical rows. Also, see Figure 3 for an expanded view of a portion of Figure 2. The genes whose expression is altered by GH treatment are obvious candidates for roles in the regulatory processes underlying the synthesis of milk.
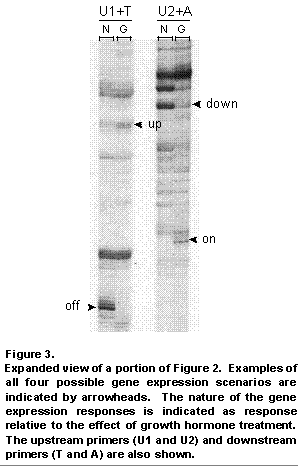
Confirmation of differential expression of the candidate genes using other molecular techniques is required to avoid selecting false positive differences. Following confirmation, and even before the identity or function of the gene has been determined, the expression of the gene can be compared in animals with different production traits, including milk composition and yield. It is expected that desirable alleles of candidate genes can be identified through that process, based on the likelihood of the presence of genetic variability (4).
It will be necessary to combine the molecular insight generated by differential display with established quantitative genetic methods in order to develop effective breeding strategies to transform existing genetic variation into the greatest possible genetic progress. The molecular uniqueness of desirable alleles would be exploited as genetic markers using the genotyping technique described below for the alleles of the k-CN and b-LG genes. Finding genes that regulate quantitative traits of economic importance almost surely will allow unforseen improvements in the productivity of dairy cattle.
Genotyping
This method is used to identify which form (allele) of a gene an animal possesses.We decided to develop the genotyping procedure using k-CN and b-LG for a very practical reason. There has been extensive research on milk protein genetic variants (6) including many studies demonstrating that certain genotypes of k-CN and b-LG are associated with specific composition and processing properties of milk (3, 8, 9, 13). Perhaps most importantly, the work has revealed which k-CN and b-LG genetic variants (alleles) are superior for cheese manufacturing. That knowledge prompted many European countries (e.g., Austria, Germany, Italy, Switzerland) to begin widespread genotyping of dairy sires and dams for milk protein genetic variants. The resulting genotype information has already been incorporated as selection criteria in dairy breeding programs. Because selection decisions depend on the economic importance of a trait, there has been more emphasis on selection of milk protein alleles in countries where a larger proportion of milk is used to make cheese (e.g., 60% in Italy).
Similar research on milk protein variants in France led to intensive selection for desirable aS1-casein (aS1-CN) alleles in goats, whose milk is mainly transformed into cheese (10). The modified breeding strategy has already reversed allelic frequencies and thereby contributed directly to improving the cheese making quality of milk.
The European initiative to exploit the genetic variability of milk proteins to enhance, through selection, the properties and cheese yield of the future milk supply represents an effective and simple beginning to a new era in dairy cattle breeding. Clearly, dairy industries in North America should adopt genotyping technology and start now to incorporate milk protein genotype information into their breeding programs. From a global perspective, not to do so is neither wise nor an option. Investing in genotyping technology today will prepare the industry for the future. With the proper tools, the dairy industry will be able to adapt to the future market environment and to remain competitive.
As mentioned previously, the development of RFLP analysis allowed the identification or detection of most molecular differences in known genes. Although its discovery represented a major advance, newer molecular methods for gene analysis are now available (7, 11, 15, 17, 18). The new methods are not only more versatile than RFLP analysis, but also overcome its major technical disadvantages, not to mention its large labor and equipment requirement, and thus high cost.
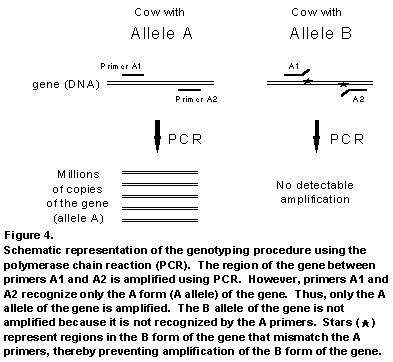 One of the main obstacles to widespread application of genotyping in the dairy industry is that it
requires a financial investment. There is an obvious need for a relatively inexpensive genotyping
technique to minimize that cost. It also follows that even with the discovery of markers for traits
with economic value, such as milk protein variants, those traits should be measurable at the
lowest possible cost. The need for an inexpensive method of genotyping many animals prompted
our work in this area and led to the development of a simple and reliable genotyping procedure
that is based on the use of state of the art molecular technique.
One of the main obstacles to widespread application of genotyping in the dairy industry is that it
requires a financial investment. There is an obvious need for a relatively inexpensive genotyping
technique to minimize that cost. It also follows that even with the discovery of markers for traits
with economic value, such as milk protein variants, those traits should be measurable at the
lowest possible cost. The need for an inexpensive method of genotyping many animals prompted
our work in this area and led to the development of a simple and reliable genotyping procedure
that is based on the use of state of the art molecular technique.
The most impressive feature of our genotyping procedure is that it is performed in a single tube. That feature makes it amenable to automation and therefore suited to commercial application (12).
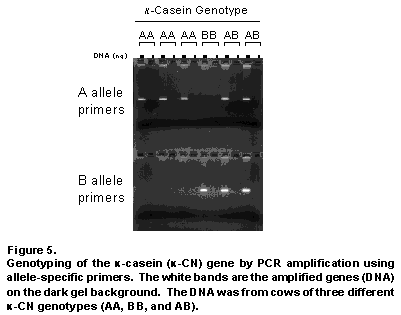
The genotyping procedure, depicted schematically in Figure 4, relies on the use of DNA primers that recognize only one specific form (allele) of the gene being evaluated. Actually, two primers are used for each allele to ensure that recognition of the allele is as specific as possible. The use of two specific primers is also essential because it allows a special protein/enzyme (DNA polymerase) to copy or amplify the part of the gene contained between the two regions recognized by the specific primers. The process of copying the gene hundreds of millions of times using PCR is what allows us to actually visualize the gene. Because we know the size of the region of DNA copied, we can confirm the success of the procedure by analyzing the copied DNA on a gel, which separates the DNA by size and compares it to other pieces of DNA of known sizes. The effectiveness of our genotyping procedure is demonstrated in Figure 5. Very little DNA is needed to perform the procedure and DNA can be obtained from blood, hair, semen, or milk.
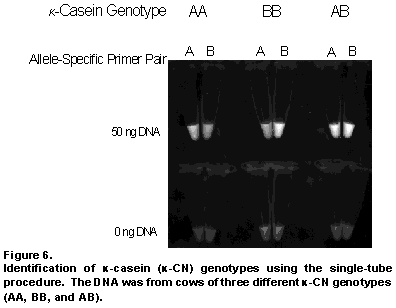
To eliminate the requirement for the procedure of gel electrophoresis to analyze the results, we transferred the gel-based procedure to a testing format that involves the use of only one tube. The principle underlying the simplified genotyping procedure is the detection of the presence of amplified DNA using a fluorescent dye. When the two allele-specific primers are in the presence of their corresponding form of the gene, then they amplify the gene and the resulting accumulated DNA combines with the fluorescent dye to fluoresce or "light up". The effectiveness of genotyping with the single-tube method is shown in Figure 6. For comparison, results are shown from three different cows, each with a different -CN genotype (AA, BB, and AB). Each tube contains DNA from one cow and one set of allele-specific primers, which recognize only one form (allele) of the -CN gene. If the cow's DNA and primers match, then the tube "lights up". Alternatively, no fluorescence is seen if the DNA and primers do not match because there is no DNA amplification.
This genotyping procedure is universally applicable to any gene. Perhaps most importantly, the use of DNA in the procedure means that all animals, regardless of sex or age, can be genotyped.
Summary
The recent development of differential display technology now allows us to solve many biological questions not previously amenable to research. Its impact in dairy science in general will surely be enormous. The most exciting aspect of using differential display in dairy genetics is the possibility of finding novel genes affecting traits of economic importance. Its use to identify genes with key roles in regulating milk synthesis and milk composition are but only two examples. We envision that searches will soon begin for genes important in many biological processes. For example, expect interest in finding genes involved in the regulatory processes underlying disease resistance, reproduction, and even feed intake. As new, important genes are discovered and characterized the resulting information can then be incorporated into dairy breeding programs through the application of large-scale genotyping. Thereby, DNA technology will contribute to improving the efficiency of production in the dairy industry.
References
1. Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts and J.D. Watson. 1989. In: Molecular Biology of the Cell. 2nd ed. Garland Publishing Inc., New York, NY. pp. 180-198.2. Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith and K. Struhl. 1994. In: Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY. pp. 15.8.1-15.8.7.
3. Bovenhuis, H., J.A.M. Van Arendonk and S. Korver. 1992. Associations between milk protein polymorphisms and milk production traits. J. Dairy Sci. 75:2549-2559.
4. Grompe, M. 1993. The rapid detection of unknown mutations in nucleic acids. Nature Genetics 5:111-117.
5. Ebert, K. M. and J.E.S. Schindler 1993. Transgenic farm animals: progress report. Theriogenology 39:121-135.
6. Jakob, E. and Z. Puhan. 1995. Implications of genetic polymorphism of milk proteins on production and processing of milk. In: bulletin of the International Dairy Federation (IDF Seminar March 28-29), Zurich, Switzerland. pp. 2-24.
7. Lee, L.G., C.R. Connell and W. Bloch. 1993. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucl. Acids Res. 21:3761-3766.
8. Lin, C.Y., A.J. McAllister, K.F. Ng-Kwai-Hang, J.F. Hayes, T.R. Batra, A.J. Lee, G.L. Roy, J.A. Vesely, J.M. Wauthy and K.A. Winter. 1989. Relationships of milk protein types to lifetime performance. J. Dairy Sci. 72:3085-3090.
9. Mao, L.I., L.G. Buttazzoni and R. Aleandri 1992. Effects of polymorphic milk protein genes on milk yield and composition traits in Holstein cattle. Acta Agric. Scand., Sect. A, Animal Sci. 42:1-7.
10. Martin, P., Ch. Leroux, Y. Amigues, M. Jans P‚rez, F. Remeuf, G. Brignon, J.-P. Furet, L. Vassal, M.-F. Mah‚, E. Manfredi, A. Jaubert, G. Ricordeau, J. Bouillon, B. Ribadeau Dumas and F. Grosclaude. 1995. Molecular diversity of the goat aS1-casein gene: Impact on casein content and cheesmaking properties of milk. In: bulletin of the International Dairy Federation (IDF Seminar March 28-29), Zurich, Switzerland. pp. 12-13.
11. Mercier, B., R. AlDaccak, A. Samaan, F. David, A. Carta, P. Cracco, O. Raguenes, F. Dufosse, C. Ferec, D. Charron, and P. Loiseau. 1994. HLA-DRB and -DBQ typing by PCR amplification using sequence-specific primers (PCR-SSP): Assessment after 1 year of routine use by three laboratories. Eur. J. Immunogenet. 21:105-110.
12. Mullis., K.B., F. Ferr‚ J and R.A. Gibbs 1994. In: The Polymerase Chain Reaction. Birkh„user, Boston, MA, pp. 182-200.
13. Ng-Kwai-Hang, K.F., H.G. Monardes and J.F. Hayes. 1990. Association between genetic polymorphism of milk proteins and production traits during three lactations. J. Dairy Sci. 73:3414-3420.
14. Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. In: Molecular cloning: A laboratory manual. 2nd ed. J. Sambrook, E. F. Fritsch, and T. Maniatis, ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
15. Skogen, B., D.B. Bellissimo, M.J. Hessner, S. Santos, R.H. Aster, P.J. Newman and J.G. McFarland. 1994. Rapid determination of platelet alloantigen genotypes by polymerase chain reaction using allele-specific primers. Transfusion: 34:955-960.
16. Soller, M. 1990. Genetic mapping of the bovine genome using deoxyribonucleic acid-level markers to identify loci affecting quantitative traits of economic importance. J. Dairy Sci. 73:2628-2646.
17. Timme, T.L. and T.C. Thompson. 1994. Rapid allelotype analysis pf p53 knockout mice. BioTechniques 17:461-463.
18. Xu., L., and B.G. Hall. 1994. SASA: A simplified, reliable method for allele-specific amplification of polymorphic sites. BioTechniques 16:44-45.